Abstract
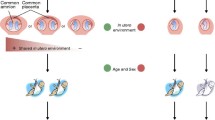
The existence of phenotypic differences between monozygotic (MZ) twins is a prime case where the relationship between genetic determinants and environmental factors is illustrated. Although virtually identical from a genetic point of view, MZ twins show a variable degree of discordance with respect to different features including susceptibility to disease. Discordance has frequently been interpreted in terms of the impact of the environment with genetics. In this sense, accumulated evidence supports the notion that environmental factors can have a long-term effect on epigenetic profiles and influence the susceptibility to disease. In relation with autoimmune diseases, the identification of DNA methylation changes in individuals who develop the disease, and the influence of inhibitors of DNA methyltransferases and histone modification enzymes in the development of autoimmunity are attracting the attention of researchers in the epigenetics field. In this context, the study of discordant MZ twins constitutes an attractive model to further investigate the epigenetic mechanisms involved in their development as well as to dissect the contribution of environmental traits. The implications of novel strategies to map epigenetic profiles and how the use of MZ twins can contribute to dissect the epigenetic component of autoimmune disease are discussed.


Similar content being viewed by others
References
MacGillivray I, Campbell DM, Thompson B (eds) (1988) Twinning and twins. Wiley, NY
Reed TE, Chandler JH (1958) Huntington’s chorea in Michigan. I. Demography and genetics. Am J Hum Genet 10:201–225
Gibbons RJ, Higgs DR (1996) The alpha-thalassemia/mental retardation syndromes. Medicine (Baltimore) 75:45–52
Greaves MF, Maia AT, Wiemels JL, Ford AM (2003) Leukemia in twins: lessons in natural history. Blood 102:2321–2333
Hrubec Z, Robinette CD (1984) The study of human twins in medical research. N Engl J Med 310:435–441
Singh SM, McDonald P, Murphy B, O’Reilly R (2004) Incidental neurodevelopmental episodes in the etiology of schizophrenia: an expanded model involving epigenetics and development. Clin Genet 65:435–440
Salvetti M, Ristori G, Bomprezzi R, Pozzilli P, Leslie RD (2000) Twins: mirrors of the immune system. Immunol Today 21:342–347
Yamagishi H, Ishii C, Maeda J, Kojima Y et al (1998) Phenotypic discordance in monozygotic twins with 22q11.2 deletion. Am J Med Genet 78:319–321
Hillebrand G, Siebert R, Simeoni E, Santer R (2000) DiGeorge syndrome with discordant phenotype in monozygotic twins. J Med Genet 37:E23
Ballestar E, Esteller M (2008) Epigenetic gene regulation in cancer. Adv Genet 61:247–267
Weksberg R, Shuman C, Caluseriu O et al (2002) Discordant KCNQ1OT1 imprinting in sets of monozygotic twins discordant for Beckwith-Wiedemann syndrome. Hum Mol Genet 11:1317–1325
Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY (1999) Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 23:185–188
Hansen RS, Wijmenga C, Luo P et al (1999) The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci U S A 96:14412–14417
Egger G, Liang G, Aparicio A, Jones PA (2004) Epigenetics in human disease and prospects for epigenetic therapy. Nature 429:457–463
Fisher AG (2002) Cellular identity and lineage choice. Nat Rev Immunol 2:977–982
Miller OJ, Schnedl W, Allen J, Erlanger BF (1974) 5-Methylcytosine localised in mammalian constitutive heterochromatin. Nature 251:636–637
Gardiner-Garden M, Frommer M (1987) CpG islands in vertebrate genomes. J Mol Biol 196:261–282
Aissani B, Bernardi G (1991) CpG islands: features and distribution in the genomes of vertebrates. Gene 106:173–183
Keshet I, Lieman-Hurwitz J, Cedar H (1986) DNA methylation affects the formation of active chromatin. Cell 44:535–543
Reik W, Collick A, Norris ML, Barton SC, Surani MA (1987) Genomic imprinting determines methylation of parental alleles in transgenic mice. Nature 328:248–251
Wolf SF, Migeon BR (1982) Studies of X chromosome DNA methylation in normal human cells. Nature 295:667–671
Ezhkova E, Pasolli HA, Parker JS et al (2009) Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136:1122–1135
Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403:41–45
Wang Y, Fischle W, Cheung W, Jacobs S, Khorasanizadeh S, Allis CD (2004) Beyond the double helix: writing and reading the histone code. Novartis Found Symp 259:3–17
Chahal SS, Matthews HR, Bradbury EM (1980) Acetylation of histone H4 and its role in chromatin structure and function. Nature 287:76–79
Rundlett SE, Carmen AA, Suka N, Turner BM, Grunstein M (1998) Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392:831–835
Santos-Rosa H, Schneider R, Bannister AJ et al (2002) Active genes are tri-methylated at K4 of histone H3. Nature 419:407–411
Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116–120
Schotta G, Lachner M, Sarma K et al (2004) A silencing pathway to induce H3–K9 and H4–K20 trimethylation at constitutive heterochromatin. Genes Dev 18:1251–1262
Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T (2002) Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep 3:39–44
de la Cruz X, Lois S, Sanchez-Molina S, Martinez-Balbas MA (2005) Do protein motifs read the histone code? BioEssays 27:164–175
Schlesinger Y, Straussman R, Keshet I et al (2007) Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 39:232–236
Widschwendter M, Fiegl H, Egle D et al (2007) Epigenetic stem cell signature in cancer. Nat Genet 39:157–158
Ballestar E, Wolffe AP (2001) Methyl-CpG-binding proteins: targeting specific gene repression. Eur J Biochem 268:1–6
Yoon HG, Chan DW, Reynolds AB, Qin J, Wong J (2003) N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol Cell 12:723–734
Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110–113
Espada J, Ballestar E, Fraga MF et al (2004) Human DNMT1 is essential to maintain the histone H3 modification pattern. J Biol Chem 279:37175–37184
Thorne JL, Campbell MJ, Turner BM (2008) Transcription factors, chromatin and cancer. Int J Biochem Cell Biol 41:164–175
McEwan IJ (2009) Nuclear receptors: one big family. Methods Mol Biol 505:3–18
Wilson VL, Jones PA (1983) DNA methylation decreases in aging but not in immortal cells. Science 220:1055–1057
Mays-Hoopes L, Chao W, Butcher HC, Huang RC (1986) Decreased methylation of the major mouse long interspersed repeated DNA during aging and in myeloma cells. Dev Genet 7:65–73
Friso S, Choi SW, Girelli D et al (2002) A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci U S A 99:5606–5611
Millar SE, Miller MW, Stevens ME, Barsh GS (1995) Expression and transgenic studies of the mouse agouti gene provide insight into the mechanisms by which mammalian coat color patterns are generated. Development 121:3223–3232
Michaud EJ, van Vugt MJ, Bultman SJ, Sweet HO, Davisson MT, Woychik RP (1994) Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes Dev 8:1463–1472
Wolff GL, Kodell RL, Moore SR, Cooney CA (1998) Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J 12:949–957
Cooney CA, Dave AA, Wolff GL (2002) Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr 132:2393S–2400S
Doherty AS, Mann MR, Tremblay KD, Bartolomei MS, Schultz RM (2000) Differential effects of culture on imprinted H19 expression in the preimplantation mouse embryo. Biol Reprod 62:1526–1535
Mann MR, Lee SS, Doherty AS et al (2004) Selective loss of imprinting in the placenta following preimplantation development in culture. Development 131:3727–3735
Lumey LH, Stein AD, Kahn HS et al (2007) Cohort profile: the Dutch Hunger Winter families study. Int J Epidemiol 36:1196–1204
Smith FM, Garfield AS, Ward A (2006) Regulation of growth and metabolism by imprinted genes. Cytogenet Genome Res 113:279–291
Cui H, Cruz-Correa M, Giardiello FM et al (2003) Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science 299:1753–1755
Heijmans BT, Kremer D, Tobi EW, Boomsma DI, Slagboom PE (2007) Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum Mol Genet 16:547–554
Heijmans BT, Tobi EW, Stein AD et al (2008) Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A 105:17046–17049
Petronis A (2001) Human morbid genetics revisited: relevance of epigenetics. Trends Genet 17:142–146
Tremolizzo L, Carboni G, Ruzicka WB et al (2002) An epigenetic mouse model for molecular and behavioral neuropathologies related to schizophrenia vulnerability. Proc Natl Acad Sci U S A 99:17095–17100
Petronis A, Gottesman II, Kan P et al (2003) Monozygotic twins exhibit numerous epigenetic differences: clues to twin discordance? Schizophr Bull 29:169–178
Mill J, Dempster E, Caspi A, Williams B, Moffitt T, Craig I (2006) Evidence for monozygotic twin (MZ) discordance in methylation level at two CpG sites in the promoter region of the catechol-O-methyltransferase (COMT) gene. Am J Med Genet B Neuropsychiatr Genet 141B:421–425
Oates NA, van Vliet J, Duffy DL et al (2006) Increased DNA methylation at the AXIN1 gene in a monozygotic twin from a pair discordant for a caudal duplication anomaly. Am J Hum Genet 79:155–162
Yamazawa K, Kagami M, Fukami M, Matsubara K, Ogata T (2008) Monozygotic female twins discordant for Silver-Russell syndrome and hypomethylation of the H19-DMR. J Hum Genet 53:950–955
Ramagopalan SV, Dyment DA, Morrison KM et al (2008) Methylation of class II transactivator gene promoter IV is not associated with susceptibility to multiple sclerosis. BMC Med Genet 9:63
Zhang AP, Yu J, Liu JX et al (2007) The DNA methylation profile within the 5′-regulatory region of DRD2 in discordant sib pairs with schizophrenia. Schizophr Res 90:97–103
Fraga MF, Ballestar E, Paz MF et al (2005) Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A 102:10604–10609
Bergem AL, Engedal K, Kringlen E (1997) The role of heredity in late-onset Alzheimer disease and vascular dementia. A twin study. Arch Gen Psychiatry 54:264–270
Kaprio J, Tuomilehto J, Koskenvuo M et al (1992) Concordance for type 1 (insulin-dependent) and type 2 (non-insulin-dependent) diabetes mellitus in a population-based cohort of twins in Finland. Diabetologia 35:1060–1067
Kaminsky ZA, Tang T, Wang SC et al (2009) DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet 41:240–245
International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN) et al (2008) Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet 40:204–210
Ueda H, Howson JM, Esposito L et al (2003) Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 423:506–511
Leslie RDG, Elliot RB (1994) Early environmental events as a cause of IDDM. Evidence and implications. Diabetes 43:843–850
Javierre BM, Esteller M, Ballestar E (2008) Epigenetic connections between autoimmune disorders and haematological malignancies. Trends Immunol 29:616–623
Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M (1990) Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum 33:1665–1673
Kaplan MJ, Lu Q, Wu A, Attwood J, Richardson B (2004) Demethylation of promoter regulatory elements contributes to perforin overexpression in CD4+ lupus T cells. J Immunol 172:3652–3661
Lu Q, Wu A, Richardson BC (2005) Demethylation of the same promoter sequence increases CD70 expression in lupus T cells and T cells treated with lupus-inducing drugs. J Immunol 174:6212–6219
Lu Q, Kaplan M, Tay D et al (2002) Demethylation of ITGAL (CD11a) regulatory sequences in systemic lupus erythematosus. Arthritis Rheum 46:1282–1291
Esteller M (2007) Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet 8:286–298
Weber M, Davies JJ, Wittig D et al (2005) Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37:853–862
Bibikova M, Lin Z, Zhou L et al (2006) High-throughput DNA methylation profiling using universal bead arrays. Genome Res 16:383–393
Taylor KH, Kramer RS, Davis JW et al (2007) Ultradeep bisulfite sequencing analysis of DNA methylation patterns in multiple gene promoters by 454 sequencing. Cancer Res 67:8511–8518
Orlando V (2000) Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem Sci 25:99–104
Weber M, Hellmann I, Stadler MB et al (2007) Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39:457–466
Barski A, Cuddapah S, Cui K et al (2007) High-resolution profiling of histone methylations in the human genome. Cell 129:823–837
Ebers GC, Sadovnick AD (1994) The role of genetic factors in multiple sclerosis susceptibility. J Neuroimmunol 54:1–17
Aho K, Koskenvuo M, Tuominen J, Kaprio J (1986) Occurrence of rheumatoid arthritis in a nationwide series of twins. J Rheumatol 13:899–902
Järvinen P, Kaprio J, Mäkitalo R, Koskenvuo M, Aho K (1992) Systemic lupus erythematosus and related systemic diseases in a nationwide twin cohort: an increased prevalence of disease in MZ twins and concordance of disease features. J Int Med 231:67–72
Greco L, Romino R, Coto I et al (2002) The first large population based twin study of coeliac disease. Gut 50:624–628
Thompson NP, Driscoll R, Pounder RE et al (1996) Genetic versus environment in IBD: results of a British twin study. BMJ 312:95
Krueger GG, Duvic M (1994) Epidemiology of psoriasis: clinical issues. J Invest Dermatol 102:14S–18S
Arnheim N, Calabrese P (2009) Understanding what determines the frequency and pattern of human germline mutations. Nat Rev Genet 10:478–488
Barros SP, Offenbacher S (2009) Epigenetics: connecting environment and genotype to phenotype and disease. J Dent Res 88:400–408
Figueiredo LM, Cross GA, Janzen CJ (2009) Epigenetic regulation in African trypanosomes: a new kid on the block. Nat Rev Microbiol 7:504–513
Hewagama A, Richardson B (2009) The genetics and epigenetics of autoimmune diseases. J Autoimmun 33:3–11
Invernizzi P (2009) Future directions in genetic for autoimmune diseases. J Autoimmun 33:1–2
Invernizzi P, Pasini S, Selmi C, Gershwin ME, Podda M (2009) Female predominance and X chromosome defects in autoimmune diseases. J Autoimmun 33:12–16
Larizza D, Calcaterra V, Martinetti M (2009) Autoimmune stigmata in Turner syndrome: when lacks an X chromosome. J Autoimmun 33:25–30
Persani L, Rossetti R, Cacciatore C, Bonomi M (2009) Primary ovarian insufficiency: X chromosome defects and autoimmunity. J Autoimmun 33:35–41
Sawalha AH, Harley JB, Scofield RH (2009) Autoimmunity and Klinefelter's syndrome: when men have two X chromosomes. J Autoimmun 33:31–34
Wells AD (2009) New insights into the molecular basis of T cell anergy: anergy factors, avoidance sensors, and epigenetic imprinting. J Immunol 182:7331–7341
Zernicka-Goetz M, Morris SA, Bruce AW (2009) Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat Rev Genet 10:467–477
Acknowledgements
EB is supported by PI081346 (FIS) grant from the Spanish Ministry of Science and Innovation (MICINN).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ballestar, E. Epigenetics Lessons from Twins: Prospects for Autoimmune Disease. Clinic Rev Allerg Immunol 39, 30–41 (2010). https://doi.org/10.1007/s12016-009-8168-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12016-009-8168-4